Florian Huber
Professor of Data Science and Visual Analytics
University of Applied Sciences Düsseldorf
Biography
Florian Huber has been a professor of Data Science and Visual Analytics at the Center for Digitalization and Digitality (ZDD) and the Department of Media at HSD since September 1st, 2021. He conducts research and teaches in the field of Data Science, with a particular focus on the use of machine learning and visualization to analyze complex data sets.
Background
Florian Huber received his diploma in Physics from the University of Leipzig and obtained his PhD in Biological Physics there in 2012, with a thesis titled “Emergent structure formation of the actin cytoskeleton.” From 2012 to 2015, he worked as a postdoctoral researcher at AMOLF (Amsterdam) and TU Delft, where he conducted biochemical experiments and computer modeling to study the self-organization of various protein structures. In 2015, Florian Huber founded the company KÄNDI in Amsterdam to develop high-quality, sustainable sweets before starting as a Data Scientist at the Netherlands eScience Center in 2018. He worked there until 2021 on numerous projects with scientists from various fields and companies, developing new methods for evaluating complex data with a strong focus on the use of machine learning. This involved working with a wide range of different data types, including mass spectra, EEG measurements, time series, sensor data, and text.
Research
Florian Huber’s research focuses on the development and application of data science algorithms for the comparative analysis of complex data. The main objective is to combine machine learning (learning from data) with domain expertise to enable meaningful comparisons between data sets or with reference data. Complex data is often transformed into so-called latent spaces, where it can be compared or modified.
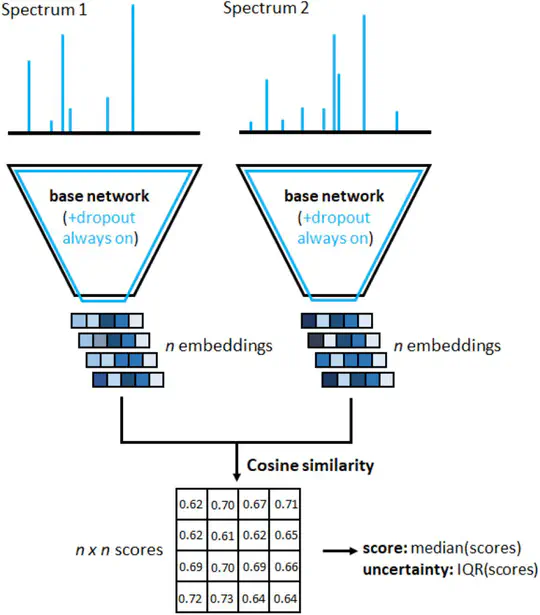
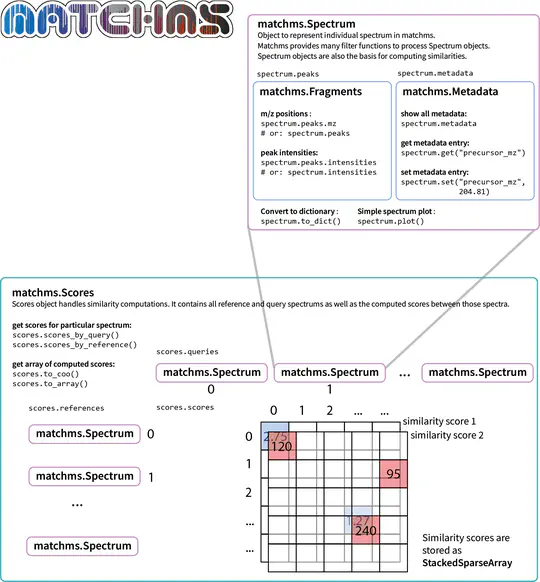
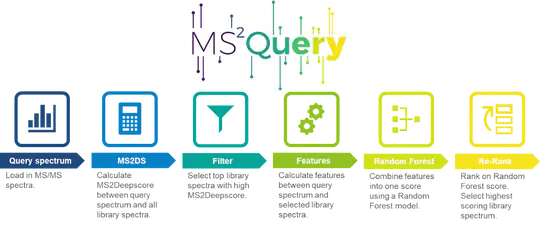
An example is the analysis of mass spectra in biology and medicine. Although these spectra are considered characteristic fingerprints for chemical substances, they cannot be attributed to a specific substance in most cases. However, the use of newly developed algorithms (machine learning, deep learning) makes it possible to reliably predict chemical similarities and locate completely unknown substances.
Other projects are currently being developed in the area of audio data (“audio data science”) as well as in the exploration and application of techniques in the area of chatbots/natural language processing (NLP).
Twitter: https://twitter.com/me_datapoint GitHub: https://github.com/florian-huber Blog: https://medium.com/@f.huber ORCID: 0000-0002-3535-9406
- Data Science
- Machine Learning (incl. Deep Learning)
- Graph-based data analysis and data visualization
- Research Software Development
- Open Science
Introduction to Data Science, 2021, 2022, 2023
University of Applied Sciences Düsseldorf
Projects